Our research projects
This section introduces current projects of the Molecular Biophysics group. Our research projects explore how molecular chaperones regulate protein assembly, linking classical aggregation kinetics with emerging concepts of phase separation and condensate biology.
-
Duration:
-
Funding source:
LCSB
-
Researchers:
-
Partners:
-
Description:
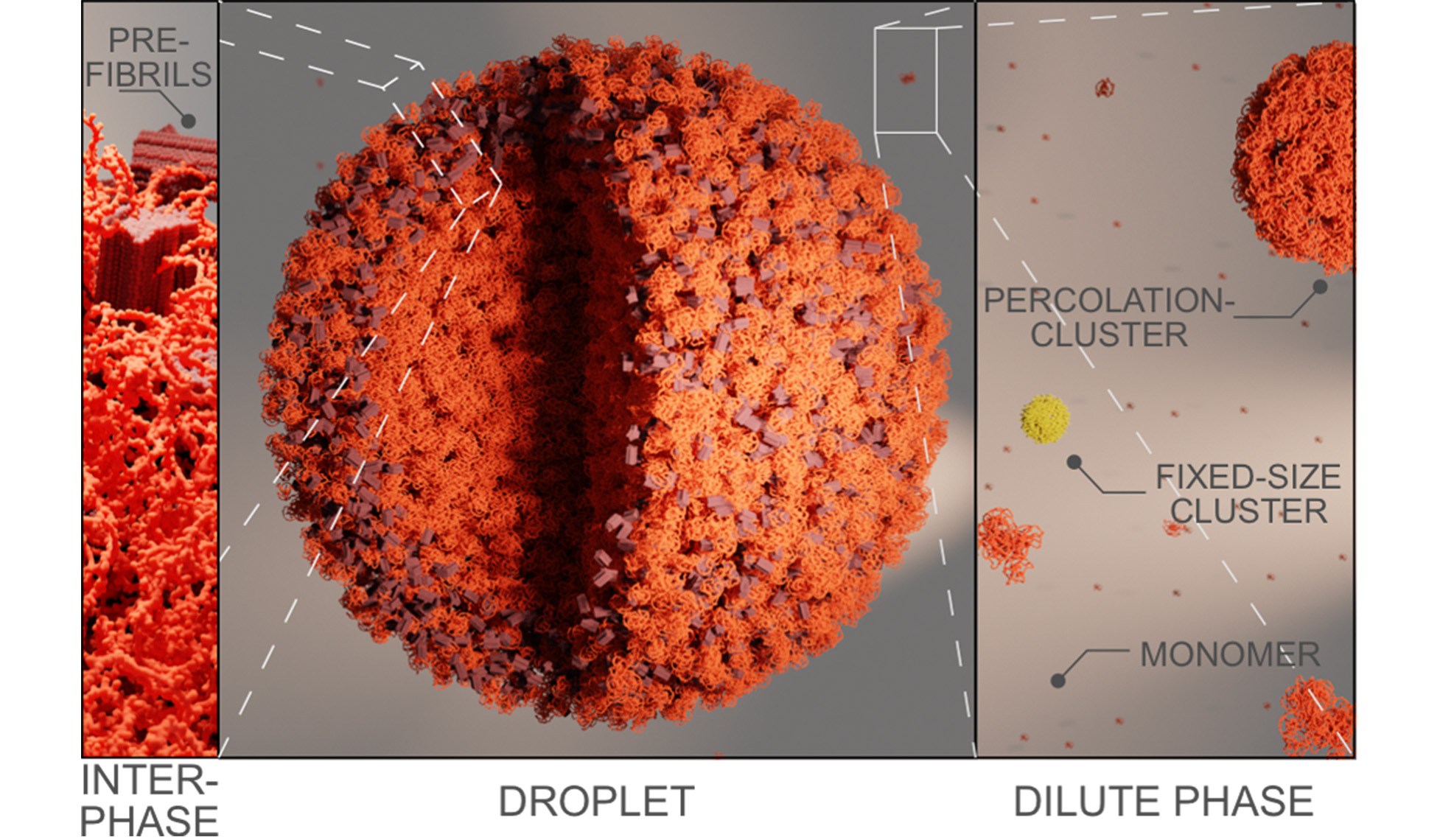
Protein aggregation underlies many neurodegenerative diseases, yet classical models of nucleation, fibril elongation, and secondary nucleation do not involve the heterogeneous protein states observed in cells. Increasing evidence points to nanometre-sized percolation clusters and liquid phase separation as potential precursors of aggregation. These assemblies are dynamic, heterogeneous, and potentially reversible, but can also convert into solid-like, pathological states. How such intermediate states are connected to classical aggregation kinetics, and how they are regulated by molecular chaperones, is largely unknown.
This project investigates how small heat shock proteins (sHSPs) control the assembly of intrinsically disordered proteins (IDPs) into clusters and condensates, and how they suppress transitions into toxic oligomers and hardened aggregates. We will dissect how dynamic sHSP oligomers engage multiple binding sites on IDPs, how they alter the surface tension and wetting behaviour of condensates, and how these interactions remodel kinetic pathways of assembly. To achieve this, we develop microfluidic diffusional sizing and electrophoresis to quantify binding equilibria in heterogeneous mixtures, employ droplet-based assays to measure wetting angles, condensate material states, and chaperone partitioning, and use Thioflavin T (ThT) fluorescence to monitor the onset and kinetics of amyloid fibril formation. Complementary fluorescence imaging will track structural transitions across scales, from nanoclusters to micrometre-sized droplets. By linking percolation cluster formation, condensate dynamics, and classical aggregation pathways, we aim to define how sHSPs safeguard the proteome and why their decline predisposes cells to sporadic neurodegeneration.
-
Project details (PDF):