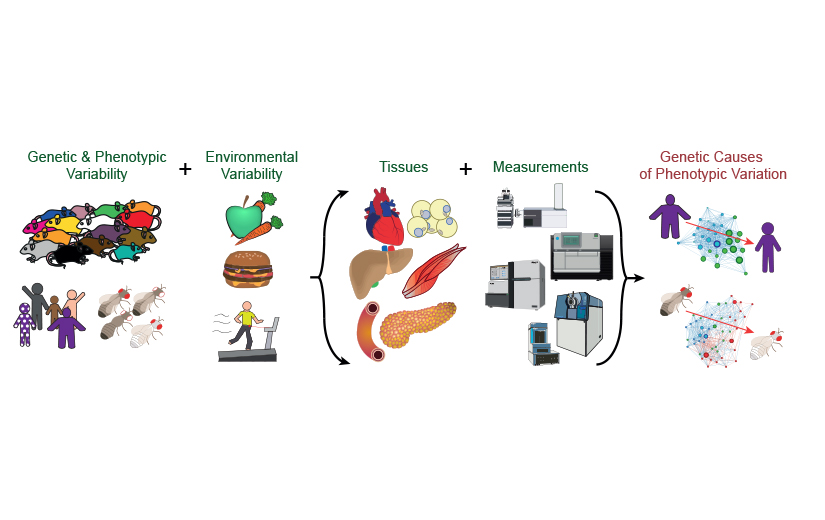
Our research projects
Our research projects are at the intersection of molecular biology, metabolism, data science, and bioinformatics. In particular, how do changes in the DNA affect expression of RNA, activity of proteins, and levels of metabolites, and how do these minute changes lead to clinical consequences?
-
Duration:
-
Funding source:
-
Researchers:
-
Partners:
-
Description:
We have previously collected tissues and data from around 700 genetically-diverse mice which have varying diets, ages, and incidences of metabolic disease. Since moving to the LCSB in 2020, we have focused on creating and analysing molecular profiles of the tissues from these individuals to determine the development of molecular perturbations preceding overt clinical manifestation of disease. This project uses causal modelling to examine which changes in regulatory networks of gene expression are preliminary drivers of metabolic disease, and which changes are downstream biomarkers of a system that has already lost homeostasis. This technique uses a time course-based analysis to determine how and which genetic and environmental factors lead to the development of disease, and then via which genes, metabolites, and regulatory mechanisms. In this project, we develop tools that allow us to move from “correlation to causality,” particularly by taking a multivariate approach where the model system is modified by multiple factors simultaneously – rather than one variable at a time as in a traditional biological approach. Candidate genes have recently been examined largely in C. elegans, but the laboratory plans to continue validation work in higher-order organisms now that this initial work has been published (Williams et al., 2021).
Long and short lifespan C. Elegans with genetic modifications
-
Project details (PDF):
-
Duration:
-
Funding source:
-
Researchers:
-
Partners:
-
Description:
Our intestines are colonised with some tens of trillions of bacterial cells which make up our “microbiota”. These bacteria can be symbiotic, enabling us to more efficiently metabolize food and to ward off disease, but they can also be detrimental, either causing disease or locking us in a negative feedback loop and inhibiting recovery. Experiments in both humans and model organisms have shown that changing our gut microbiome can change disease phenotypes, proving that microbiota can have causal effects on the host, and highlighting their potential utility in medical treatments. However, due to the huge diversity of the microbiome, much remains unknown about the cause and effect of specific changes in its composition. In our research, we examine how the gut microbiome in the cecum changes over the lifespan of a mouse. We can thus determine when the gut microbiome changes in response to differences in genetics, diet, and time, and by examining other tissues in the mice, we can see how the gut microbiome communicates with the host organism. This will allow us to define not only which parts of the gut microbiome are beneficial or detrimental, but also which changes in gene expression – at either the microbe or host level – are active.
-
Project details (PDF):
-
Duration:
-
Funding source:
-
Researchers:
-
Partners:
-
Description:
It is a common truism that exercise aids in fitness, and that certain diets have negative health effects, but there is a tremendous range in how different people respond to similar exercise and dietary regimens. We have previously examined the natural propensity of a large population of mice to the development of metabolic disease under different dietary states, and the propensity of mice to exercise when given access to suitable equipment. At the LCSB, we have started a project to merge these two prior findings. Do mice that are predisposed to obesity and diabetes, but which enjoy exercise when given the opportunity, still develop metabolic disease, and to what extent? We have just established a long-term experiment – roughly one year – where we will follow these mice over the first third of their natural lifespan, where they will eat and exercise to their own volition to determine to what extent the ability to exercise can ameliorate (or even prevent) metabolic disorders. Should these mice be able to largely mitigate the negative effects of a diet if they were living a sedentary lifestyle, we will investigate further to determine the contribution of behavioral traits (e.g. desire to exercise) versus core metabolic changes (e.g. mitochondrial morphology).

Two identical mouse sisters, one on a low fat the other on a high fat diet.
-
Project details (PDF):