Our research
The main interests of the Computational Biology group revolve around stem cell research, regenerative medicine, cell engineering and ageing. To that end we focus on developing tools and models at a multi-scale level.
Stem cell research and regenerative medicine
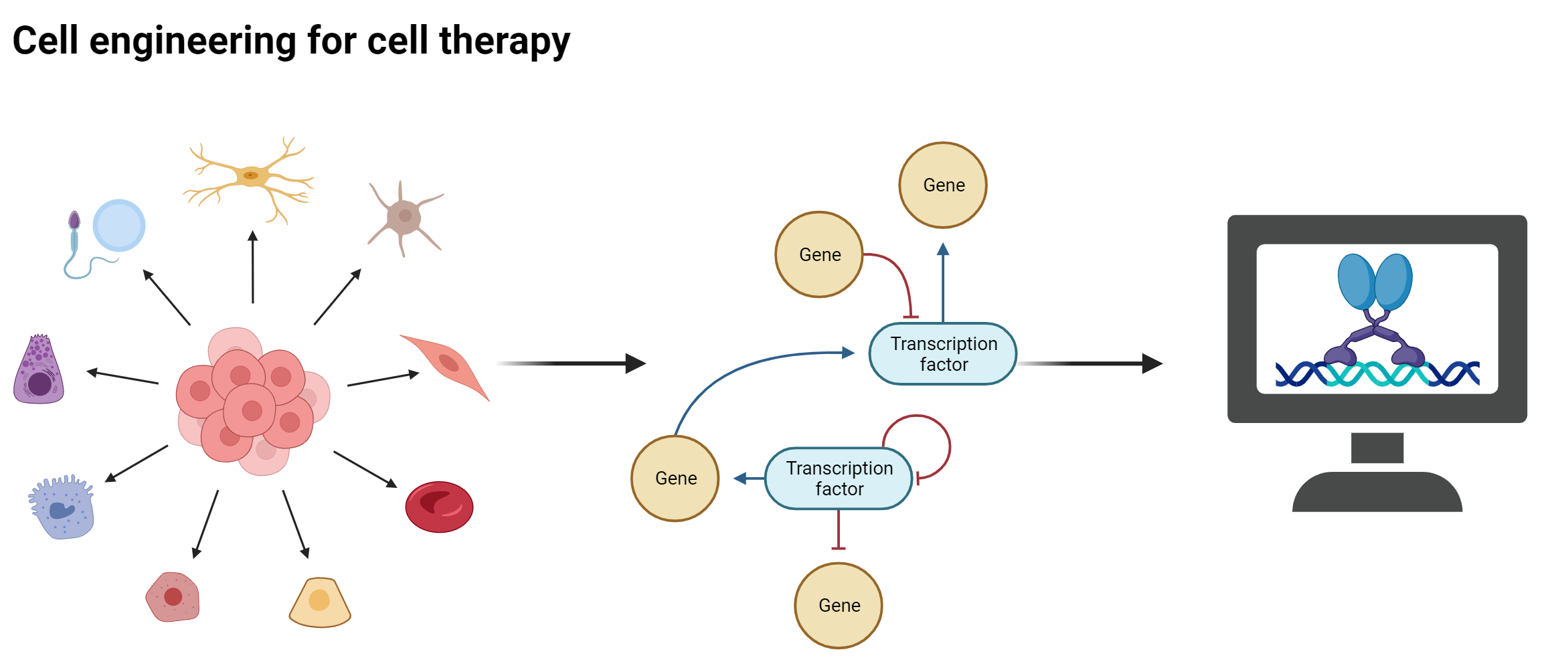
Cell therapy, which relies on the transplantation of healthy cells to replace the function of diseased or damaged cells, holds great promise for treating various diseases and conditions. However, one of the major limitations of this approach is the in vitro generation of functionally specific cell sub-populations for transplantation. We have developed a computational platform to guide cell engineering for cell therapy. This platform relies on a single cell gene regulatory network model to identify optimal sets of conversion factors to generate specific cell subpopulations. We are currently applying this methodology to projects relevant for cell therapies including the in vitro generation of corneal limbus stem cells from cultured epithelial keratinocytes. The derived experimental protocol will be further optimized and used for treating patients who have lost their corneal limbus stem cells due to injury or burn. Further, we will apply our platform to produce corticospinal tract neuron progenitors in vitro for transplantation in cases of spinal cord injuries. Finally, we have generated predictions for the efficient conversion of cardiac right ventricular cells into left counterparts, which are currently being experimentally validated. The outcome of this project will be essential for designing strategies to treat patients with cardiovascular diseases.
Ageing and rejuvenation
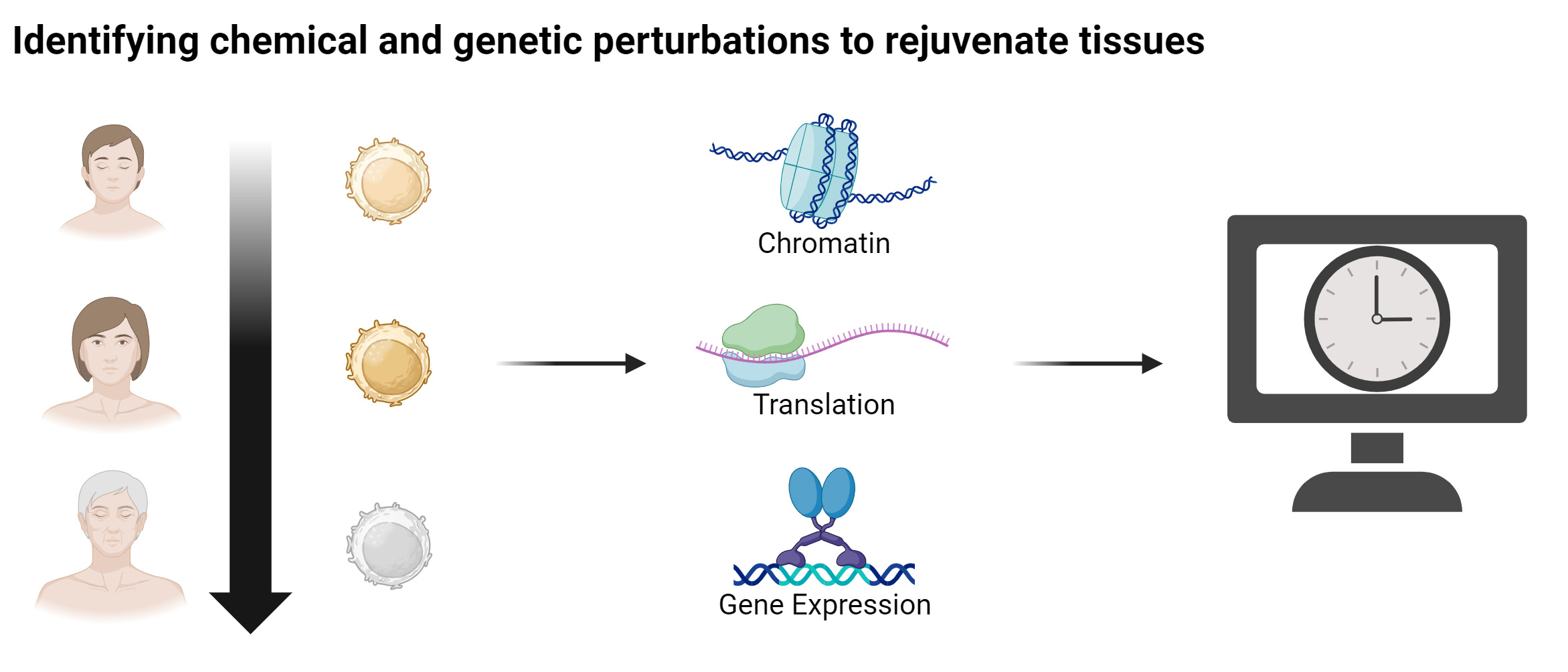
The quantification of the biological age of cells yields great promises for accelerating the discovery of novel rejuvenation strategies. In this context, we have developed the first multi-tissue RNA clock that measures the biological, rather than chronological, age of cells from their transcriptional profiles by evaluating key cellular processes. Moreover, we have also implemented a fibroblast-specific RNA clock to predict genes that can act as rejuvenation factors for human fibroblasts. We also employ gene network-based approaches to identify key molecules and pathways for cellular rejuvenation. In this context, we have identified cocktails of transcription factors for rejuvenating different cell types, including hematopoietic stem cells and hepatocytes.
Brain rejuvenation to counteract neurodegeneration
By interrogating transcriptional changes in the old human brain compared to its young counterpart, we have identified key signalling pathways and molecules that are dysregulated during brain aging. Moreover, using a computational tool recently developed in our lab, we have predicted cocktails of chemical compounds that can potentially target these dysregulated pathways to rejuvenate the brain. Experimental validation of these predictions is currently being carried out in human cells and in animal models.
Considering that ageing is an important risk factor of neurodegenerative diseases, our next goal is to apply this approach to a mouse Alzheimer’s and Parkinson’s disease model as a therapeutic strategy to counteract disease pathology.
Disease modelling
Modulating the cytokine storm in inflammatory diseases
Dysregulations in the inflammatory response of the body could progress toward a hyperinflammatory condition associated with increased severity and mortality. In particular, in the case of COVID-19 we built a comprehensive cell-cell interaction map of the immune response in patients with mild and severe symptoms to identify positive feedback loops amplifying the immune response, which led to the discovery of two novel therapeutic targets. Further, we will apply a similar strategy to modulate hyperinflammation in the systemic inflammatory response syndrome (SIRS), which is a complex disorder with nonspecific symptoms. Currently, we lack targeted therapy for SIRS not only because triggers are often unknown, but also because host responses to triggers may vary. In this regard, we collected PBMCs from a cohort of patients in order to identify common repurposable drugs targeting key molecules to reduce inflammation. We are a part of an EU-funded consortium in which experiments will be conducted to validate the computational predictions and provide mechanistic insights into patient treatments.
Increasing the differentiation capacity of stem cells in congenital disorders
Congenital disorders affect an estimated 6% of babies worldwide resulting in hundreds of thousands of associated deaths. In this regard, mutations in specific genes can lead to abnormal organogenesis characterized by an impaired differentiation potential of progenitor cells. We have developed a network-based computational method to predict lineage-specifying transcription factors that are malfunctioning in these disorders. In particular, following this approach we identified key cell fate determinants whose failure disrupt cardiac development, providing a framework to investigate congenital heart defects. Moreover, we applied this method to one of the forms of mitochondrial DNA-associated Leigh syndrome (MILS) characterized by an impaired differentiation capacity of neural progenitor cells. Our predictions identified cell fate determinants in these cells whose upregulation increases neurogenesis as well as neuronal morphogenesis, and thus can be used as a therapeutic target in this disorder. We are part of an EU-funded consortium project in which our computational predictions are being validated with the goal of identifying repurposable compounds for treating MILS.
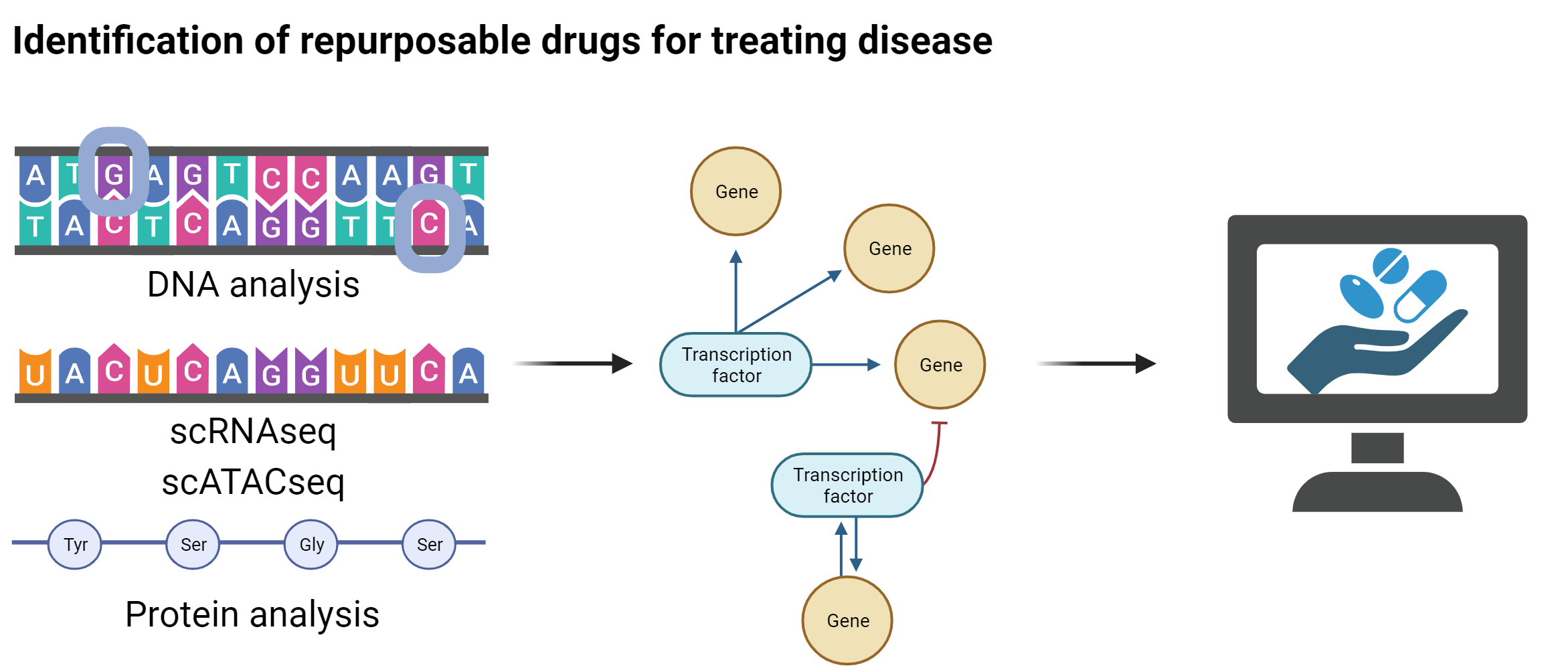
Tools
This section introduces different computational tools developed by the Computational Biology group.
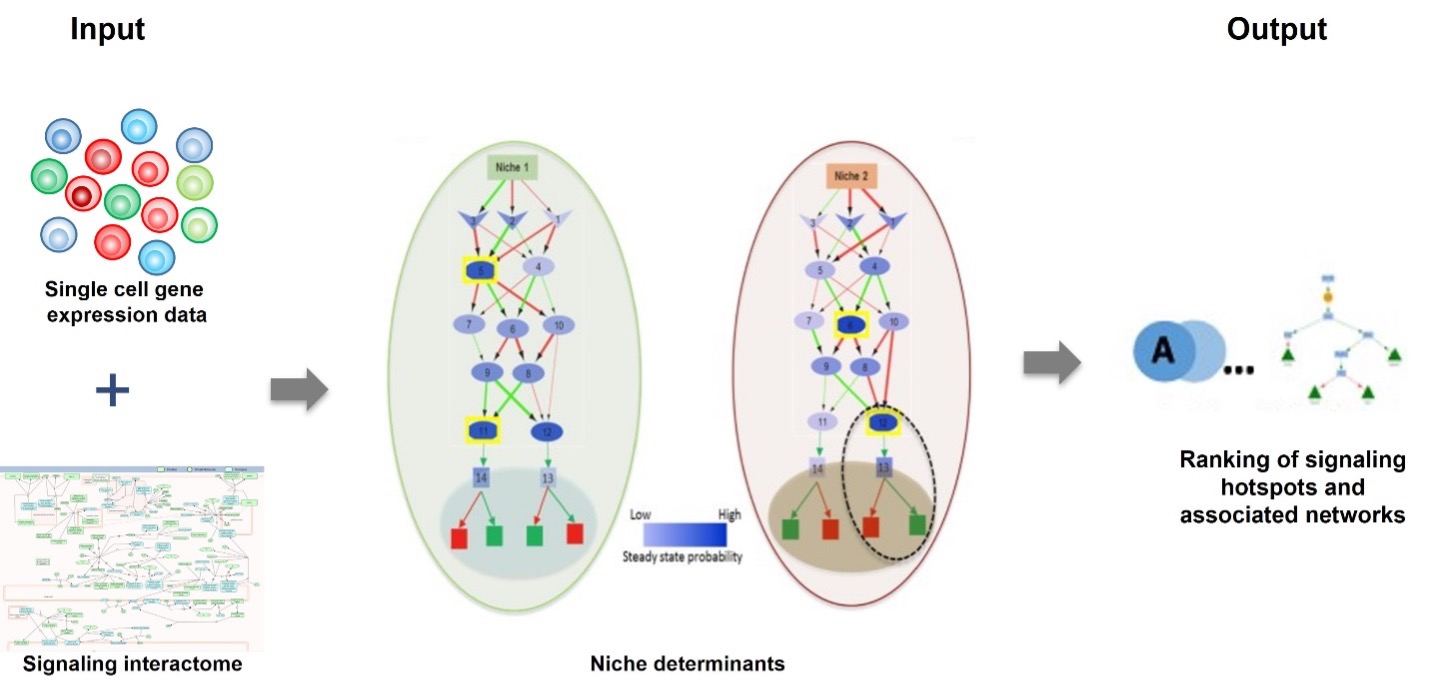
SigHotSpotter is a computational tool designed to identify crucial regulatory elements—often termed “hotspots”—in sustained signaling pathways. Through targeted analysis of these regulatory elements, SigHotSpotter provides a robust framework for manipulating cellular behaviors. SigHotSpotter is available both as a web-server and as stand-alone software.
Article : https://doi.org/10.1093/bioinformatics/btz827
The web application is available at https://sighotspotter.lcsb.uni.lu/
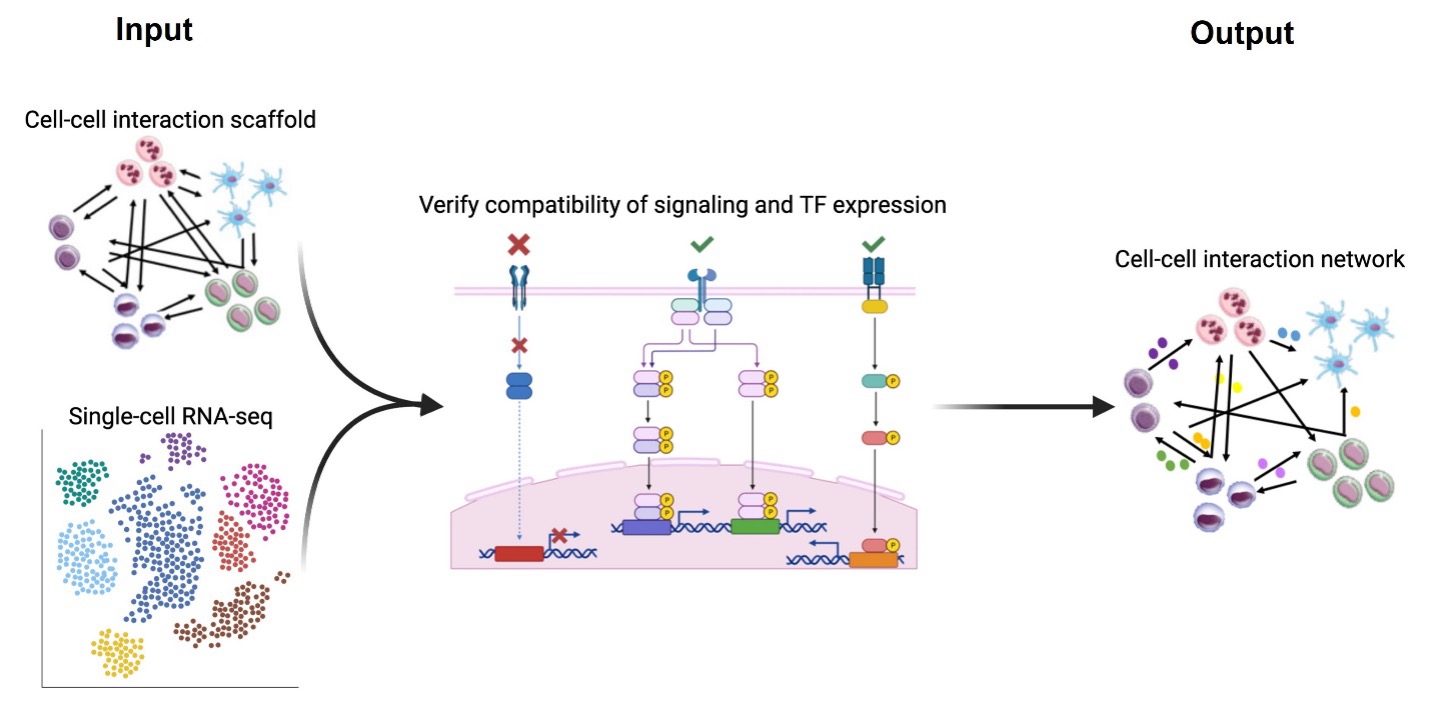
InterCom is a computational tool that infers receptor/ligand-mediated cell-to cell interactions. It extends SigHotSpotter by adding curated information about experimentally validated ligand-receptor interactions. By doing this, it is able to determine which are the receptors that are active and as a consequence infer the receptor-ligand communication between different types of cells in the dataset.
The R package information is available at https://github.com/saschajung/Intercom
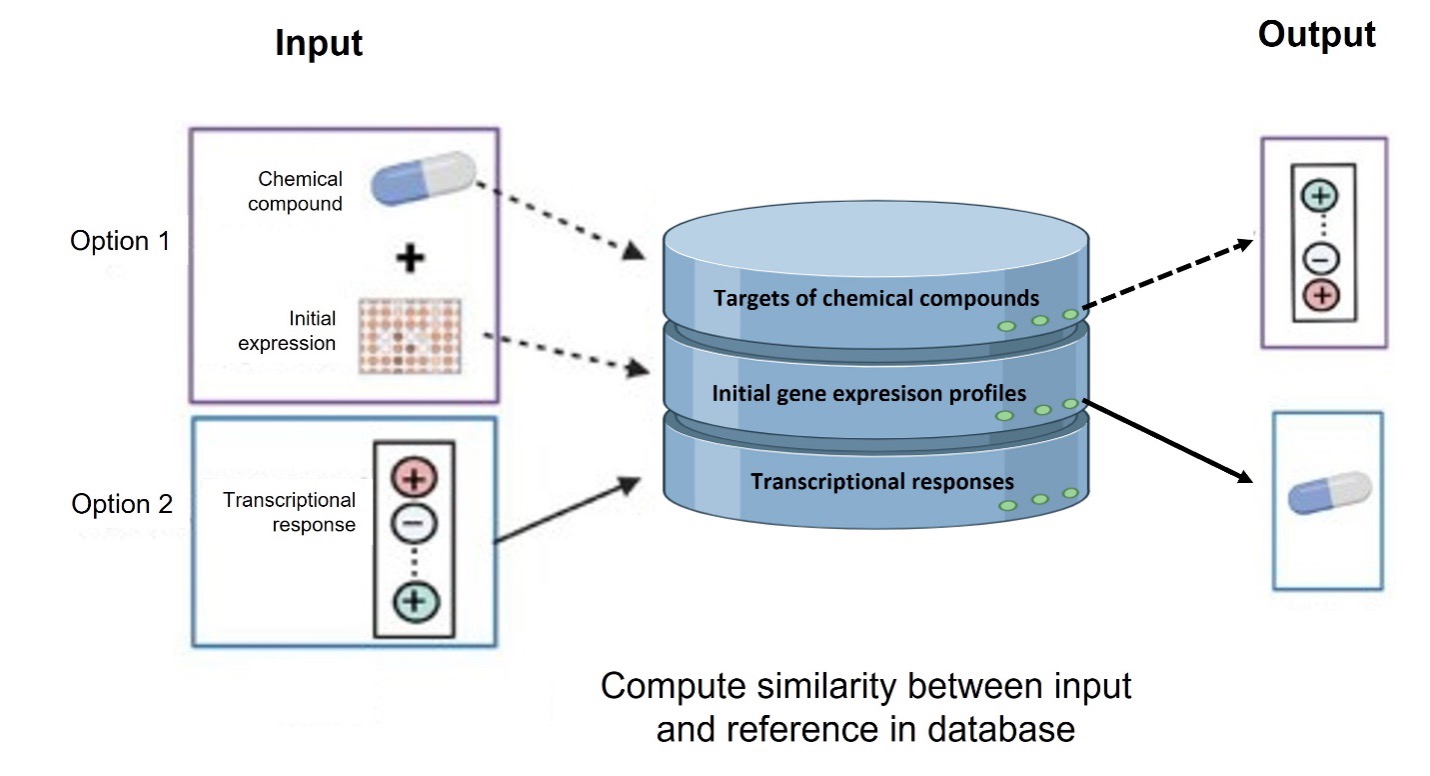
ChemPert is a specialized database and computational tool aimed at predicting the transcriptional responses of non-cancer cells to various perturbagens, including drugs, small molecules, and protein ligands. It allows both to predict the transcription factors that are likely to respond to a given perturbagen (top-down), and to obtain the best perturbagen candidates to affect a given set of target transcription factors. Accessible through a user-friendly web interface, ChemPert serves as a valuable resource for both experimental and computational researchers.
Article : https ://doi.org/10.1093/nar/gkac862
The web application is available at : https://chempert.uni.lu/
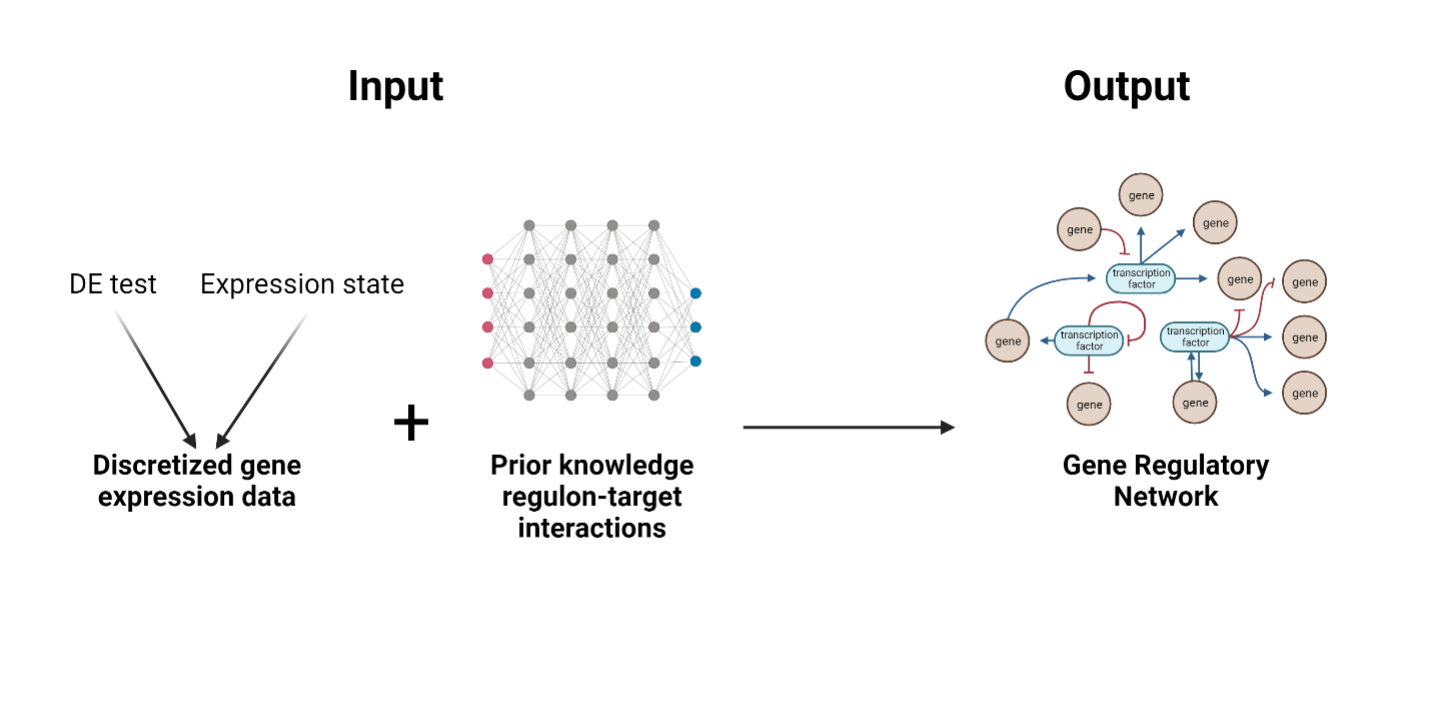
GRNopt is a specialized computational module that forms an integral part of the SeeSawPred workflow. It is designed to construct gene regulatory networks (GRNs) based on two inputs. First, it needs discretized gene expression data, which may come from a single expression state or a differential gene expression test. Second it needs a previous knowledge network of regulon-target interactions from experimental data. The output presents a finely inferred GRN, capturing the relationships between activating and inhibiting sources and their respective targets.
The R package information is available at : https://git-r3lab.uni.lu/CBG/GRNOptR
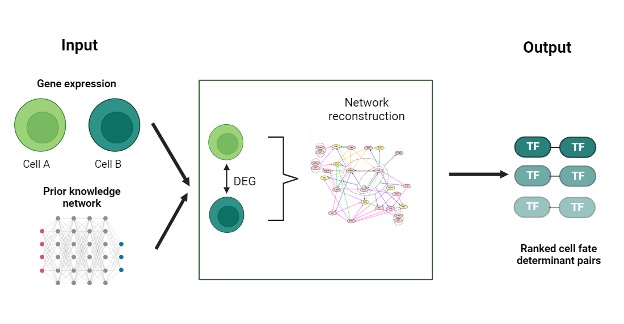
SeesawPred is a web application designed to identify cell-fate determinants, specifically transcription factors (Tfs), that guide cellular differentiation. Unlike many existing tools, SeesawPred allows for user-provided gene expression data and doesn’t depend on pre-compiled reference datasets. This makes it uniquely suited for exploring novel differentiation protocols. Built on a gene regulatory network (GRN) model, SeesawPred has been validated about both human and mouse cell differentiation examples.
Article : https://doi.org/10.1038/s41598-018-31688-9
The web application is available at : https://seesaw.lcsb.uni.lu/webapp2/
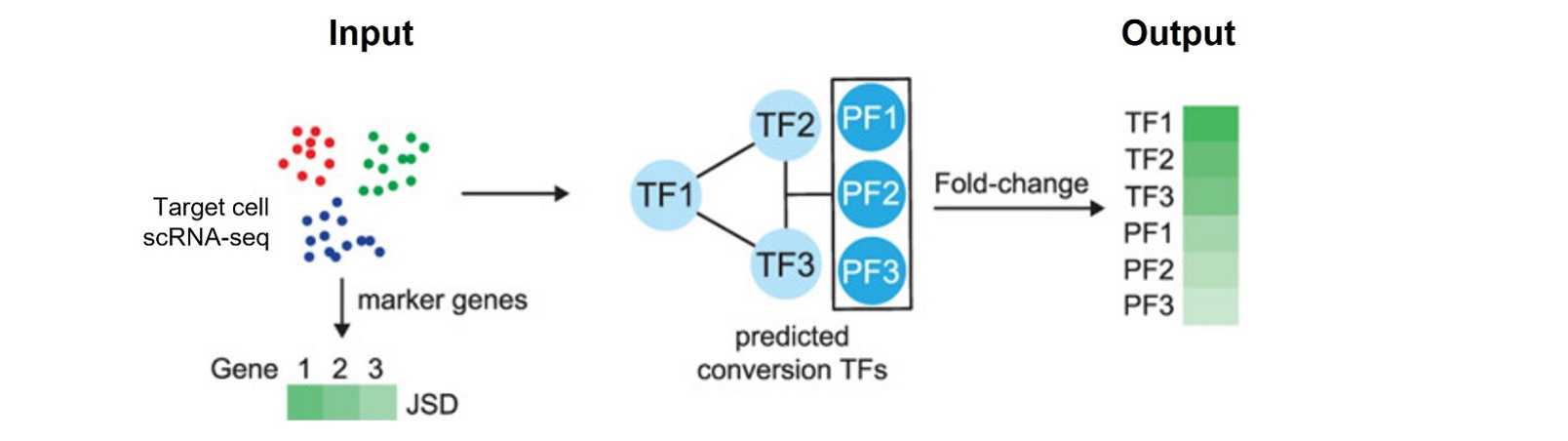
TranSynW is a web application designed to facilitate the generation of desired cell types through cell conversion. Utilizing scRNA-seq data, it identifies and prioritizes transcription factors (TFs), focusing on pioneering factors crucial for chromatin opening, for user-specified cell populations. Additionally, TransSynW also suggests marker genes for evaluating the success of cell conversion experiments. TransSynW does not require specialized computational skills or resources, making it a valuable asset for experimentalists in stem cell research and regenerative medicine.
Article : https://doi.org/10.1002/sctm.20-0227
The web application is available at : https://transsynw.lcsb.uni.lu/
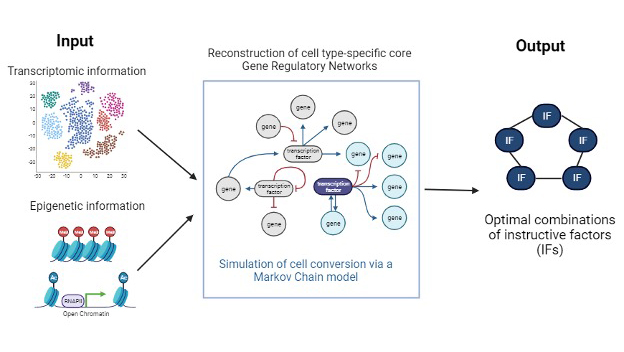
IRENE is a computational tool developed to enhance the efficiency of cellular conversion technologies. It integrates a stochastic gene regulatory network model to prioritize instructive factors (IFs) for cellular conversions. The tool is unique in its ability to align the transcriptional and epigenetic landscapes of converted cells with their target cell types, thereby optimizing conversion rates.
Article : https://doi.org/10.1038/s41467-021-21801-4
IRENE’s workflow information is available at : https://github.com/saschajung/IRENE
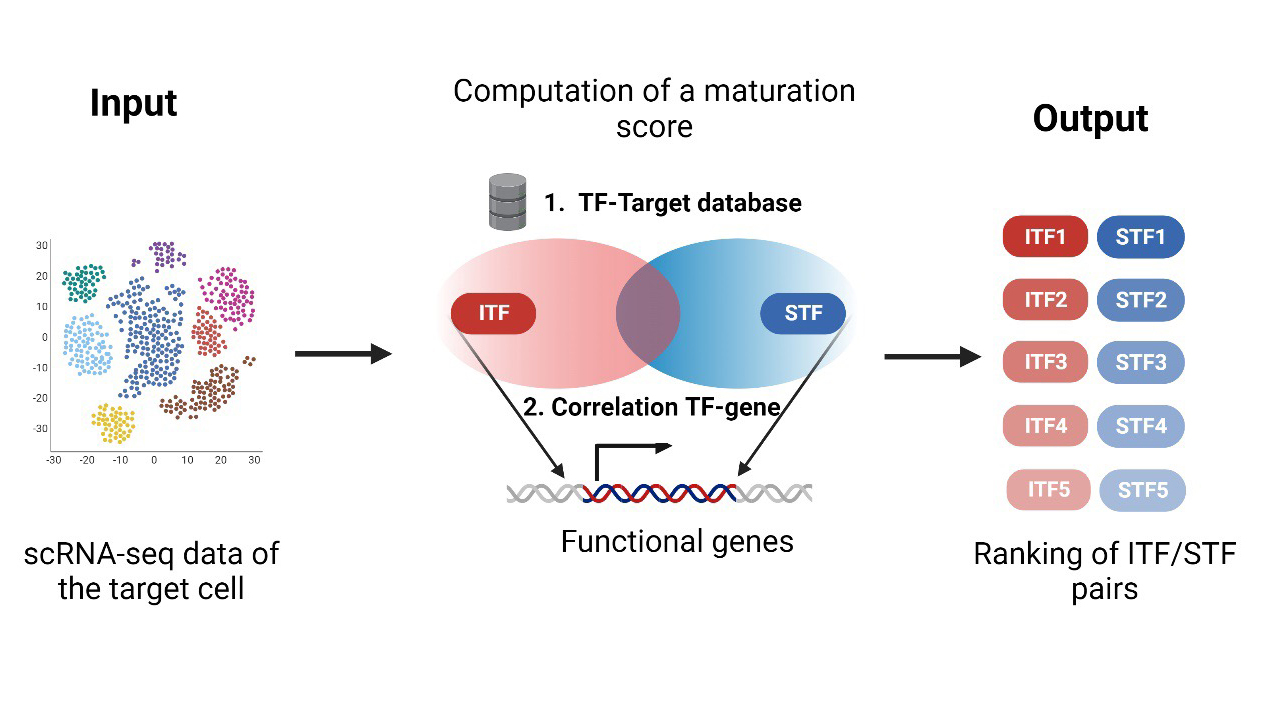
SinCMat is the first computational tool designed to facilitate the generation of fully functional and mature engineered cells. While existing computational approaches aim at predicting transcription factors (TFs) for cell differentiation/reprogramming, no method currently exist that specifically considers function cell maturation processes. Using scRNA-seq data of the target cell, SinCMat is based on a model of cell maturation that integrates identity and environment components and predicts pairs of TFs that co-target genes driving functional maturation. The web application contains the tool, SinCMatDB, a database of experimentally validated maturation cues, and an atlas of maturation TFs for mouse and human.
Article : https://doi.org/10.1016/j.stemcr.2023.12.006
The web application is available at : https://sincmat.lcsb.uni.lu/