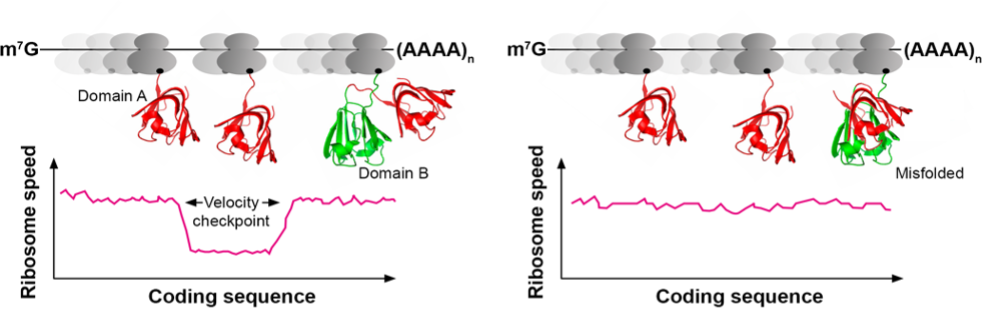
Ribosomes adjust their speed during translation. At the velocity checkpoint, they slow down to give the nascent chain enough time to find its native fold. Mutations that alter ribosome speed can lead to protein misfolding. Importantly, this can be the case for both non-synonymous and synonymous silent mutations.
Our research projects
Our research projects integrate translatomics, interactomics, visual proteomics and machine learning based structure analysis to unravel the impact of ribosome speed on co-translational folding in healthy and diseased cells.
-
Duration:
2025-2028
-
Funding source:
LCSB
-
Researchers:
-
Partners:
-
Description:
In this project we will use induced pluripotent stem cells and differentiate them into neural stem cells and neurons (1). Then we will identify ribosome pause sites using (kinetic) ribosome profiling (2). This method provides a snapshot of ribosome footprints along mRNAs. These pause sites will be aligned with the protein domain structure to identify velocity checkpoints where the ribosome slows down to facilitate co-translational folding. The experiments will be complemented by structure analysis using AlphaFold and structure probing using limited proteolysis.
- O’Neill AC et al. Spatial centrosome proteome of human neural cells uncovers disease-relevant heterogeneity. Science. 2022 Jun 17;376(6599):eabf9088. doi: 10.1126/science.abf9088. Epub 2022 Jun 17. PMID: 35709258.
- Ingolia NT et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009 Apr 10;324(5924):218-23. doi: 10.1126/science.1168978.
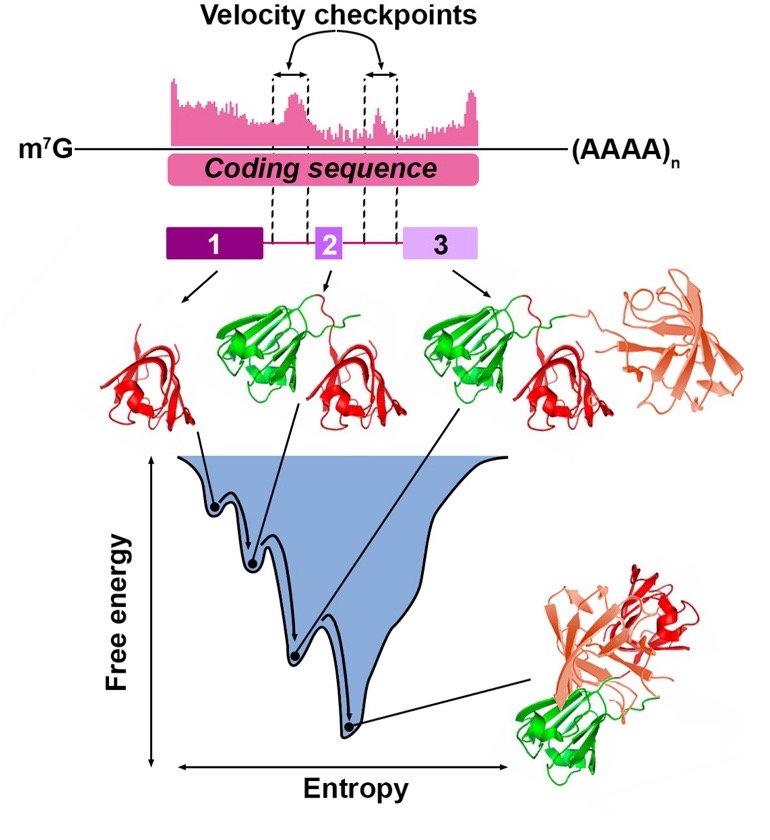
We will analyse footprints of actively translating ribosomes to identify pause sites. These sites will be aligned with the domain structure of the encoded proteins to identify velocity checkpoints that will allow reconstruction of co-translational folding trajectories.
-
Project details (PDF):
-
Duration:
2025-2027
-
Funding source:
Cure SMA
-
Researchers:
-
Partners:
Dr. Gabriella Viero, Prof. Marina Boido, Dr. Tobias Straub
-
Description:
Spinal muscular atrophy (SMA) is a neurodegenerative disease characterised by the loss of motor neurons in the spinal cord (1). Newly developed therapies for SMA have added years to the life expectancy of severely affected patients. As a result, new symptoms have emerged in treated patients, such as intellectual deficits (2), suggesting a complex pathological impact of the disease.
In this project, funded by Cure SMA (https://www.curesma.org/), we will investigate the role of the Survival Motor Neuron (SMN) protein in regulating the epitranscriptome and translatome required for higher cognition (3). To this end, we will integrate iCLIP data with ribosome profiling and SLAM-seq using SMN-deficient neurons and mouse brains.
- Mercuri E et al. Spinal muscular atrophy. Nat Rev Dis Primers. 2022 Aug 4;8(1):52. doi: 10.1038/s41572-022-00380-8. PMID: 35927425.
- Baranello G; Neurodevelopment in SMA Working Group. The emerging spectrum of neurodevelopmental comorbidities in early-onset Spinal Muscular Atrophy. Eur J Paediatr Neurol. 2024 Jan;48:67-68. doi: 10.1016/j.ejpn.2023.11.006.
Shi H et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018 Nov;563(7730):249-253. doi: 10.1038/s41586-018-0666-1.
-
Project details (PDF):
